Congratulations to PhD candidate, Hanyu, for successfully presenting his 3rd-year seminar titled “Impacts of Synthesis Conditions on Membrane Protein Incorporation into Nanodiscs”! Great job, Hanyu!
Manik presents his 3rd-year seminar!
Congratulations to PhD candidate, Manik, for successfully presenting his 3rd-year seminar titled “Derivation of Pediatric Sepsis Biomarker Signatures Using Photonic Microring Resonators”! Great job, Manik!
Lindsay presents her 3rd-year seminar!
Congratulations to PhD candidate, Lindsay, for successfully presenting her 3rd-year seminar titled “A Dynamic Phase Grating Approach to Probing Content in Microfluidic Droplet Trains”! Great job, Lindsay!
Ayush presents his 3rd-year seminar!
Congratulations to PhD candidate, Ayush, for successfully presenting his 3rd-year seminar titled “Development of a Droplet Microfluidics Platform for the Rapid Detection of Electrolytes”! Great job, Ayush!
Manik awarded summer fellowship!
Congratulations to 3rd-year student Manik Reddy who was awarded the Spring/Summer Carpenter, D. C. Memorial Fellowship through the chemistry department! Way to go!
Congratulations to all the departmental fellowship awardees!
New Publication! Congratulations to Krista, Cole, Heather, and Manik on their work in PLOS One!
Congratulations to Dr. Krista Meserve on her recent first author publication in PLOS One. Congratulations also to Dr. Cole Chapman, Dr. Heather Robison, and 3rd-year PhD Candidate Manik Reddy for their contributions to this work.
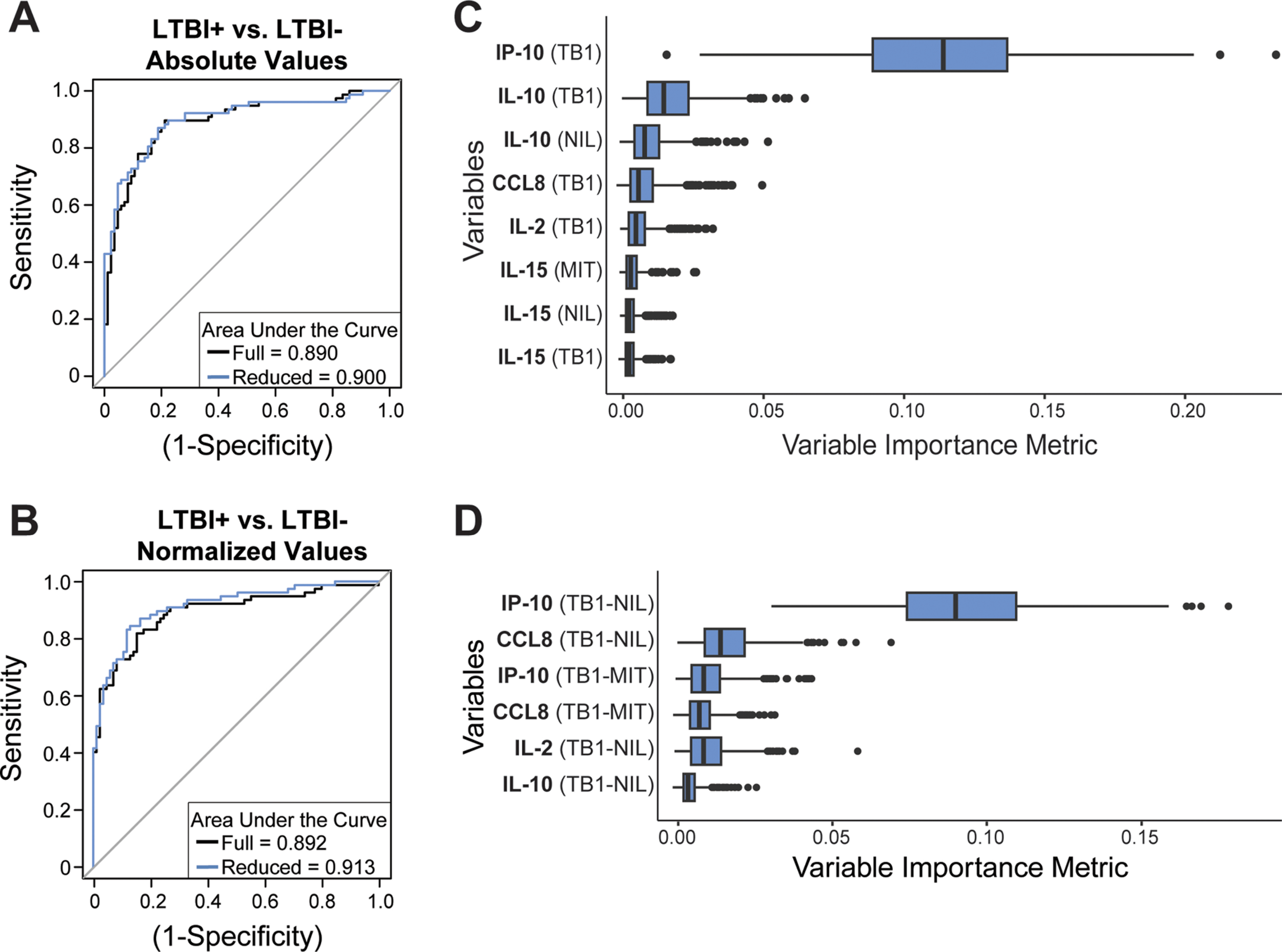
Multiplexed cytokine profiling identifies diagnostic signatures for latent tuberculosis and reactivation risk stratification
Abstract: Active tuberculosis (TB) is caused by Mycobacterium tuberculosis (Mtb) bacteria and is characterized by multiple phases of infection, leading to difficulty in diagnosing and treating infected individuals. Patients with latent tuberculosis infection (LTBI) can reactivate to the active phase of infection following perturbation of the dynamic bacterial and immunological equilibrium, which can potentially lead to further Mtb transmission. However, current diagnostics often lack specificity for LTBI and do not inform on TB reactivation risk. We hypothesized that immune profiling readily available QuantiFERON-TB Gold Plus (QFT) plasma supernatant samples could improve LTBI diagnostics and infer risk of TB reactivation. We applied a whispering gallery mode, silicon photonic microring resonator biosensor platform to simultaneously quantify thirteen host proteins in QFT-stimulated plasma samples. Using machine learning algorithms, the biomarker concentrations were used to classify patients into relevant clinical bins for LTBI diagnosis or TB reactivation risk based on clinical evaluation at the time of sample collection. We report accuracies of over 90% for stratifying LTBI + from LTBI– patients and accuracies reaching over 80% for classifying LTBI + patients as being at high or low risk of reactivation. Our results suggest a strong reliance on a subset of biomarkers from the multiplexed assay, specifically IP-10 for LTBI classification and IL-10 and IL-2 for TB reactivation risk assessment. Taken together, this work introduces a 45-minute, multiplexed biomarker assay into the current TB diagnostic workflow and provides a single method capable of classifying patients by LTBI status and TB reactivation risk, which has the potential to improve diagnostic evaluations, personalize treatment and management plans, and optimize targeted preventive strategies in Mtb infections.
Anusha passes her gateway exam!!
Congratulations to 2nd-year PhD candidate, Anusha Vajrala, for completing her gateway exam at the beginning of the month! Here’s to successful microring applications to infectious diseases and mosquito-borne flaviviruses in the years to come!
Congrats, Anusha, for completing this significant milestone in your graduate school journey! We are all so proud of you!
Pittcon Conference 2025
The 2025 Pittcon conference and exposition was held on March 1st-5th in Boston, MA. This event brought together analytical scientists in academia and industry with a keynote lecture presented by Professor Cato T. Laurencin from the University of Connecticut. Third year students Ayush Chitrakar, Manik Reddy and Hanyu Zheng represented the Bailey lab with poster presentations!
Professor Ryan Bailey was the organizer for the session “Analyst at 150: The Longest-serving Measurement Science Journal Continues to Shape the Field.” During the session, he gave a talk titled “Multiplexed Biomarker Analysis to Improve Diagnostics of Infections and Infectious Diseases.”
Congratulations to all the presenters!
Ryan speaks at Ain Shams University in Cairo, Egypt!
Ryan traveled to Cairo, Egypt at the end of January to present a talk titled “Multiplexed Biomarker Analysis to Improve Diagnostics of Infections and Infectious Diseases” to the Department of Pharmaceutical Chemistry at Ain Shams University!
New Publication! Congratulations to Marina on her work in Analytical Chemistry!
Congratulations to Dr. Marina Sarcinella on her recent first author publication in Analytical Chemistry alongside Associate Chair for Research and Professor of Chemistry, Dr. Brandon Ruotolo, and Hobart H Willard Distinguished University Professor of Chemistry and Pharmacology, Dr. Robert Kennedy. Congrats, also, to Sam Edgcombe for the work she contributed to this publication!
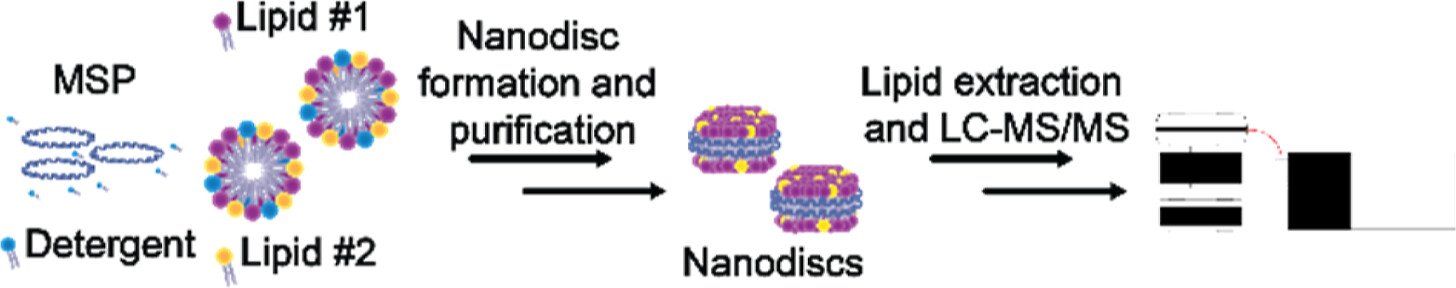
Lipid Curvature and Fluidity Influence Lipid Incorporation Disparities in Nanodiscs
Abstract: Nanodiscs have become a popular membrane mimetic system offering a well-defined bilayer environment to stabilize membrane proteins for in vitro analyses using a range of analytical methods; however, lipid compositions common to their deployment are simplistic and often fail to model native membrane complexity. Furthermore, there has been a general lack of rigorous analytical and biophysical characterization of nanodiscs comprising more than one lipid. To address these challenges, we coupled a nanodisc formation and purification workflow with targeted LC–MS/MS analysis to quantify lipids in nanodiscs made with different compositions. We screened lipids with a variety of headgroups and acyl chains and found that lipids did not always incorporate into nanodiscs at expected levels. Disparities in lipid incorporation were found to increase upon the addition of lipids known to induce curvature or rigidity to the membrane. Additionally, we found that adding just one additional type of lipid to nanodiscs changes the particle diameter and dispersity compared to nanodiscs containing a single lipid. We also formed and characterized nanodiscs using a complex starting composition inspired by the endoplasmic reticulum membrane and observed native-like cholesterol dynamics that modulated the lipid fluidity in the model bilayer system. Taken together, this work serves as a foundation for understanding nonstoichiometric lipid incorporation into nanodiscs and provides a basis for more thorough nanodisc characterization and quality control, which is critical to ensure multilipid nanodiscs synthesized accurately model the biological system of interest, enabling robust characterization of how the lipid landscape affects membrane protein structure and activity.