Multiplexed microRNA expression profiling by combined asymmetric PCR and label-free detection using silicon photonic sensor arrays
ABSTRACT
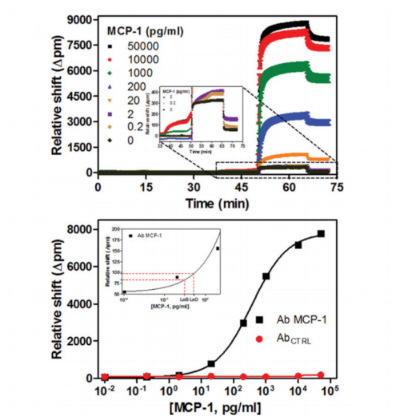
Analysis methods based upon the quantitative, real-time polymerase chain reaction are extremely powerful; however, they face intrinsic limitations in terms of target multiplexing. In contrast, silicon photonic microring resonators represent a modularly multiplexable sensor array technology that is well-suited to the analysis of targeted biomarker panels. In this manuscript we employ an asymmetric polymerase chain reaction approach to selectively amplify copies of cDNAs generated from targeted miRNAs before multiplexed, label-free quantitation through hybridization to microring resonator arrays pre-functionalized with capture sequences. This method, which shows applicability to low input amounts and a large dynamic range, was demonstrated for the simultaneous detection of eight microRNA targets from twenty primary brain tumor samples with expression profiles in good agreement with literature precedent. Click here to read the manuscript.